P1: Estimation of bi-allelic mutation rates at Y-STRs – Call for a Collaborative Project
Organizers
Nádia Pinto1, 2, 3, Leonor Gusmão4, António Amorim1,2,5
- Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Porto, Portugal
- Instituto de Patologia e Imunologia Molecular da Universidade do Porto (IPATIMUP), Porto, Portugal
- Center of Mathematics of the University of Porto, Porto, Portugal
- Laboratório de Diagnóstico por DNA (LDD), Universidade do Estado do Rio de Janeiro (UERJ), Rio de Janeiro, Brazil
- Faculdade de Ciências, Universidade do Porto, Porto, Portugal
Description
Estimating (overall) mutation rates through to the simple proportioning of cases showing Mendelian incompatibilities is a formally sound and statistically robust method only in the case of Y-chromosomal transmission, where it is unambiguous which allele at the father originated which allele at the son. Several works were already published presenting estimates for Y-allelic mutation rates, but few present the data in a format which would allow the accurate inclusion of the relevant parameters in the statistical calculations, as already highlighted at N. Pinto, L. Gusmão, A. Amorim, Mutation and mutation rates at Y chromosome specific Short tandem Repeat Polymorphisms (STRs): A reappraisal, Forensic Sci. Int. Genet. 9 (2014), 20-24. Indeed, from a formal point of view, the mutational parameter required in a likelihood ratio is bi-allelic specific (i.e. the pertinent mutation rate is the allelic transition observed), for whose estimation the absolute number of “non-mutated” transitions is also required. Thus, the primer goal of this call is to estimate bi-allelic mutations rates by asking collaboration from authors who have already developed work for estimating Y-STRs mutation rates in a Y-haplotypic framework. Moreover, large datasets in a haplotype framework will allow also to determine if: (a) the haplotype background influences the (bi-allelic) mutation rates, and (b) the probability of occurrence of a mutation at a locus influences the probability of occurrence of other mutations at other loci, in the same gametogenesis.
Requested data and format
We ask for collaborations which allow us to collect Y-haplotypic information in a large scale, as complete as possible, exclusively from father-son duos and including all tested transmissions (both mutated and non-mutated). If possible, information such as father’s age at the gametogenesis and a brief characterization of the population (preferably using YHRD nomenclature) should be provided. Please do not include ‘repeated fathers’, i.e. one father and more than one son. The requested format is a simple Excel worksheet as follows:
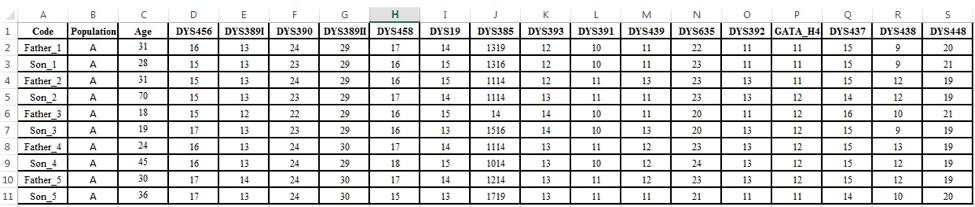
Deadline
Expired at April 29th, 2016.


